
EpiTYPER DNA Methylation analysis
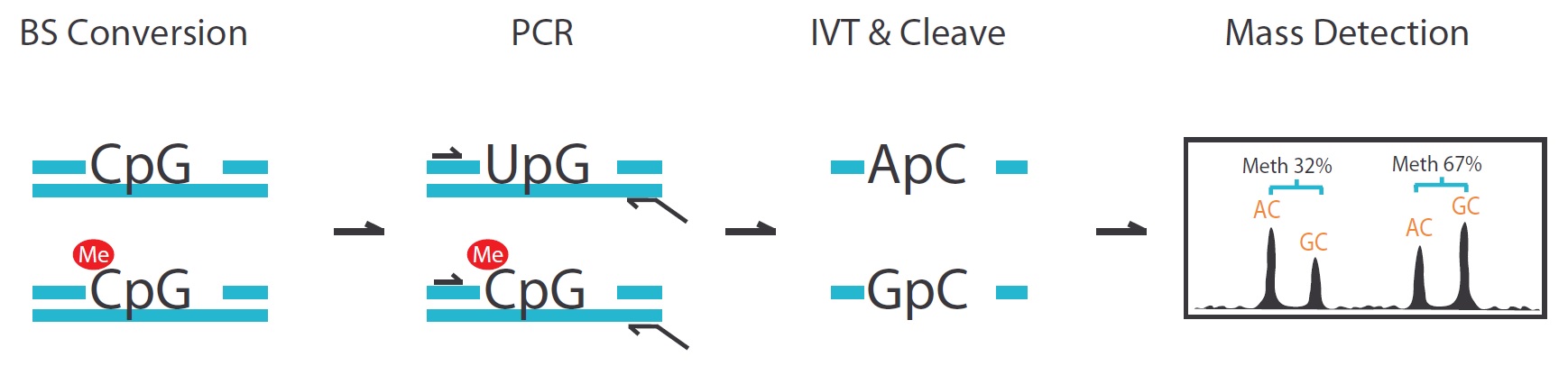
There are four steps in the EpiTYPER DNA Methylation analysis, including bisulfite conversion, PCR, MassCLEAVE (T7-IVT and Base-specific RNA cleavage) and Mass detection.
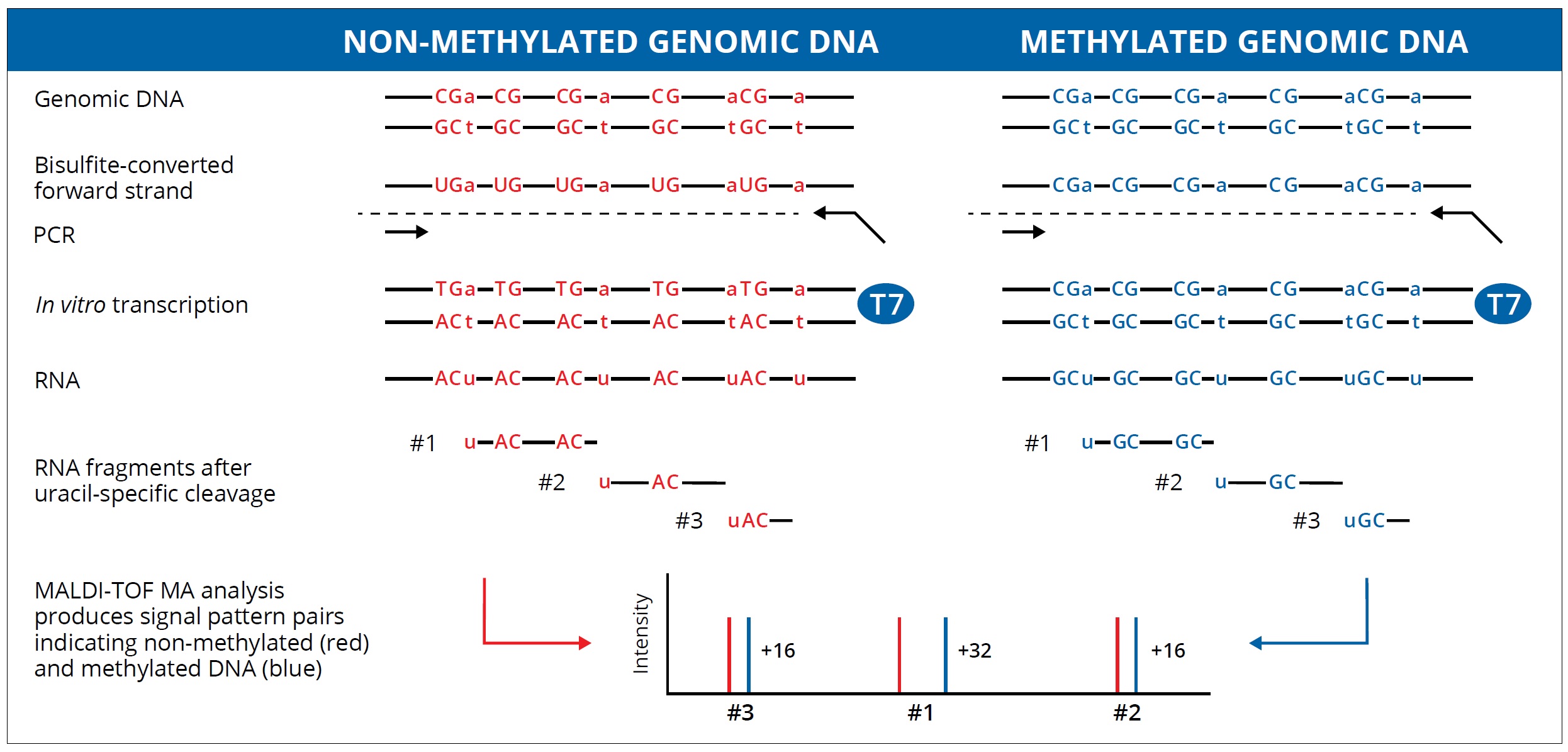
- The unmethylated cytosines in the DNA sequence are converted to uridines after the bisulfite treatment.
- Then T7 RNA polymerase will bind to the T7 RNA polymerase promoter sequence on the reverse PCR primer and perform in vitro transcription and transfer the CTP into dCTP.
- In the meantime, the single strand RNA product will be cleaved by RNase A during the MassCLEAVE process.
- The whole sequence will be cleaved to tens of fragments generally. The fragment that contained CpG sites will have two kinds of molecular weight (GC or AG) because of the methylation status. There are 16 Daltons of discrepancy between the methylated and unmethylated sequence. We can report the methylation ratio data base on the signal area.
Advantages:
- MALDI-TOF technology offers high accuracy of quantitative methylation data
- Data CV< 5%, quantitative linearity between 5% to 95%
- Up to 600bp DNA fragment can be analysed in a single reaction
- No sequencing, limited hands-on time
- Cost-effective and high sample throughput
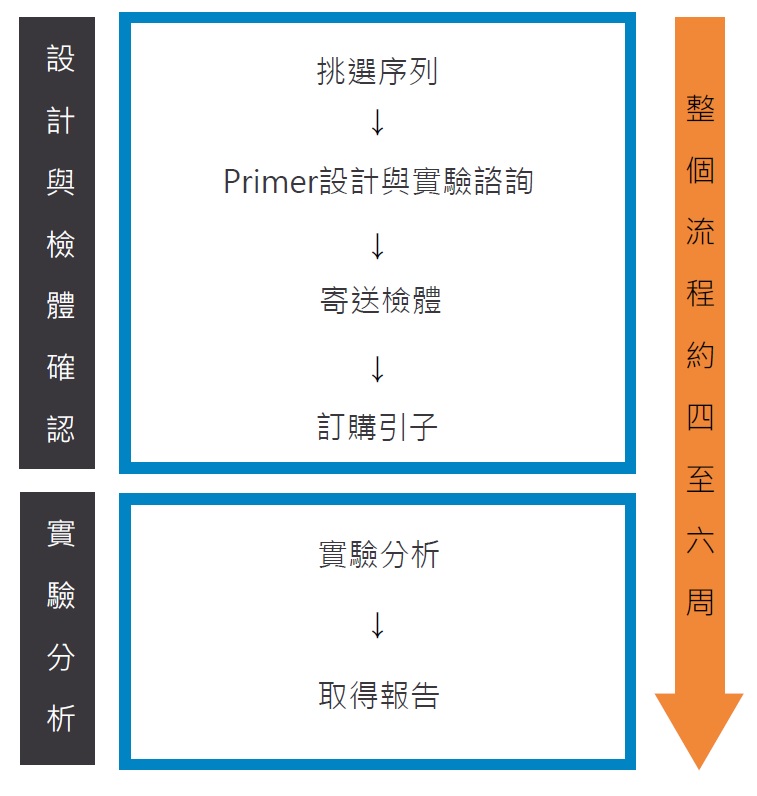
Methylation analysis Service Workflow:
- Prepare sequence list.
- We offer the primer design report
- Do the bisulfite treatment, run the electrophoresis, and send us the picture of the bisulfite PCR products
- Prepare the samples and sample list as the following format
- The genomic DNA samples should be loaded in the 96 well plate or PCR strip tubes
- At least 24 reactions is needed (reaction No. = sample No.* amplicon No.).
- At least 10 µl per bisulfite PCR product is needed.
Sample Requirements:
Accepted formats : Genomic DNA/Bisulfite PCR product
Applicable samples : Blood/FFPE gDNA
Minimum analysis unit : 24 reactions
Total requirement : 2μg gDNA/10μl Bisulfite PCR product
